Absolute Quantification of Protein and mRNA Abundances Demonstrate Variability in Gene-Specific Translation Efficiency in Yeast
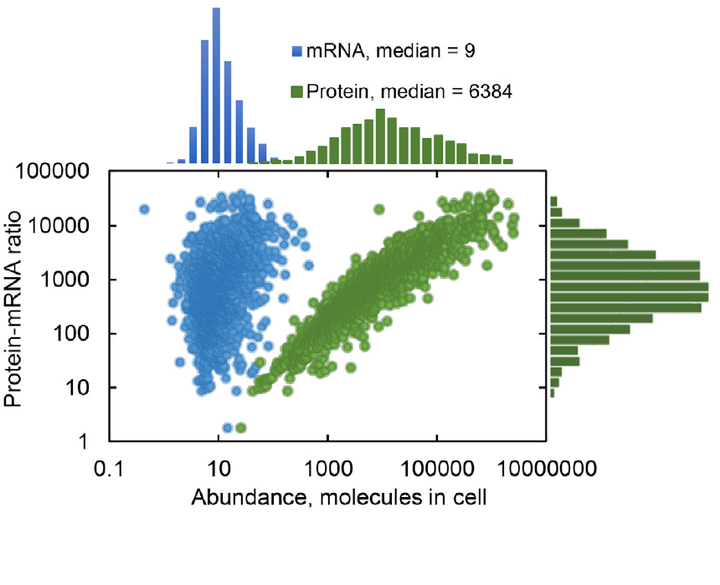 Distribution of the number of different mRNAs and proteins in yeast. Taken from the original publication: https://www.doi.org/10.1016/j.cels.2017.03.003
Distribution of the number of different mRNAs and proteins in yeast. Taken from the original publication: https://www.doi.org/10.1016/j.cels.2017.03.003Abstract
In order to get an overview of the physiological adaptation to stress in S. cerevisiae, in this study we measured absolute abundances (i.e. number of copies per cell) of over 5000 mRNAs and 2000 proteins, under 10 different conditions of stress (heat stress, osmotic stress and ethanol stress). We also estimated degradation rates for over 1000 proteins, in order to compute variability in translational efficiency across different processes in the cell. This multi-layered dataset has proven extremely helpful in my research, and I keep coming back to it whenever I want to validate methods or unveil more yeast biology.
Type
Publication
Cell Systems